Linear Amplicon Sequencing
Linear Amplicon Sequencing
Please input each sample name on a separate line. Please review our sample labeling and packing instructions if this is your first order with us (especially if submitting plates).
Couldn't load pickup availability
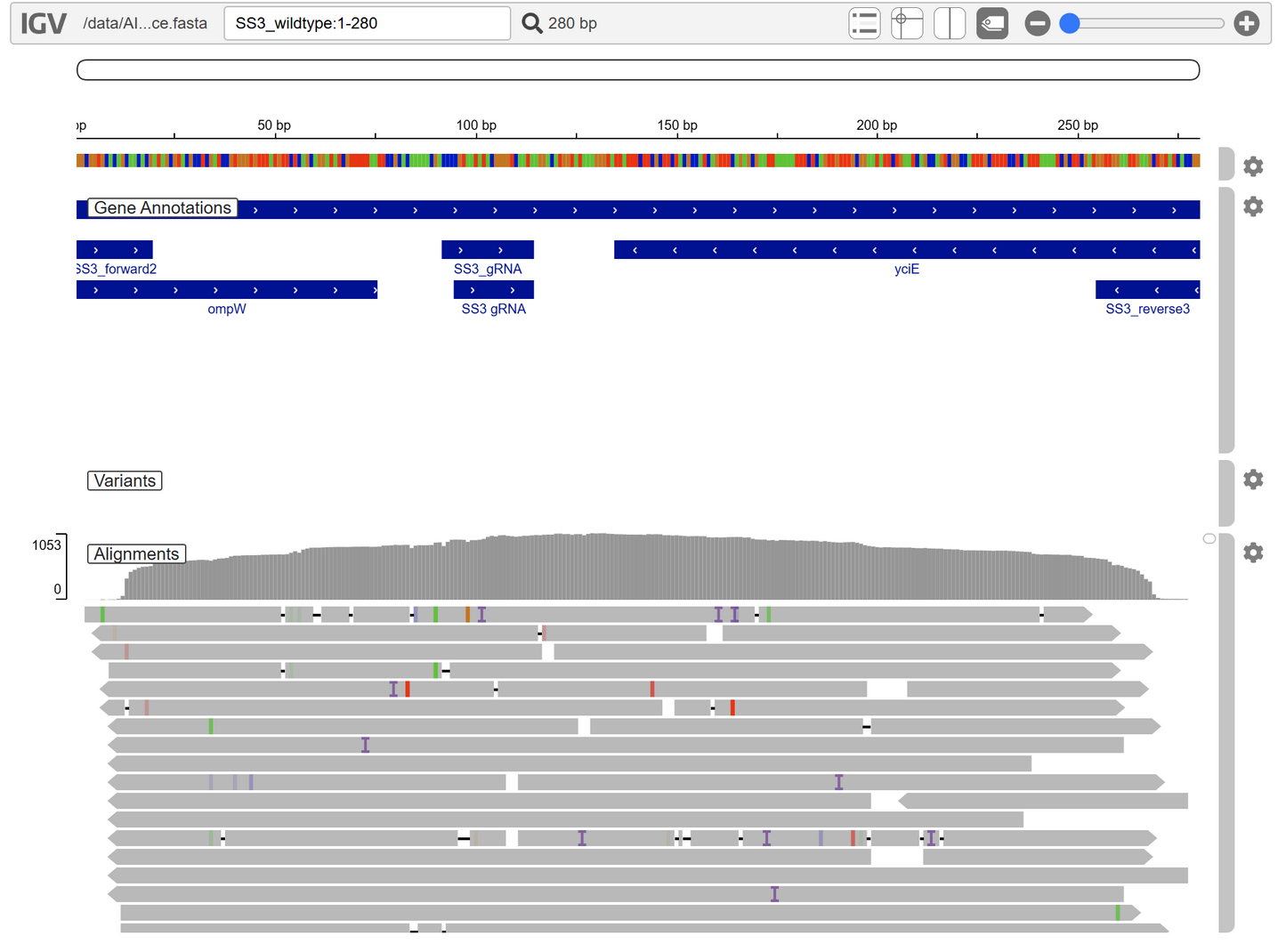
Click here to see an example with SS3 amplicon validation
✅ No reference sequence is required to build the consensus.
✅ If you provide a reference, we’ll align your results and highlight any unwanted mutations.
How to ensure high quality amplicon results?
Please make sure that you provide high enough concentration (between 20-200 ng/μL). We highly encourage to measure the concentration using fluorometric quantitation such as Qubit. Nanodrop can provide incorrect results that often overestimates the concentration.
Do you offer full coverage of the amplicons?
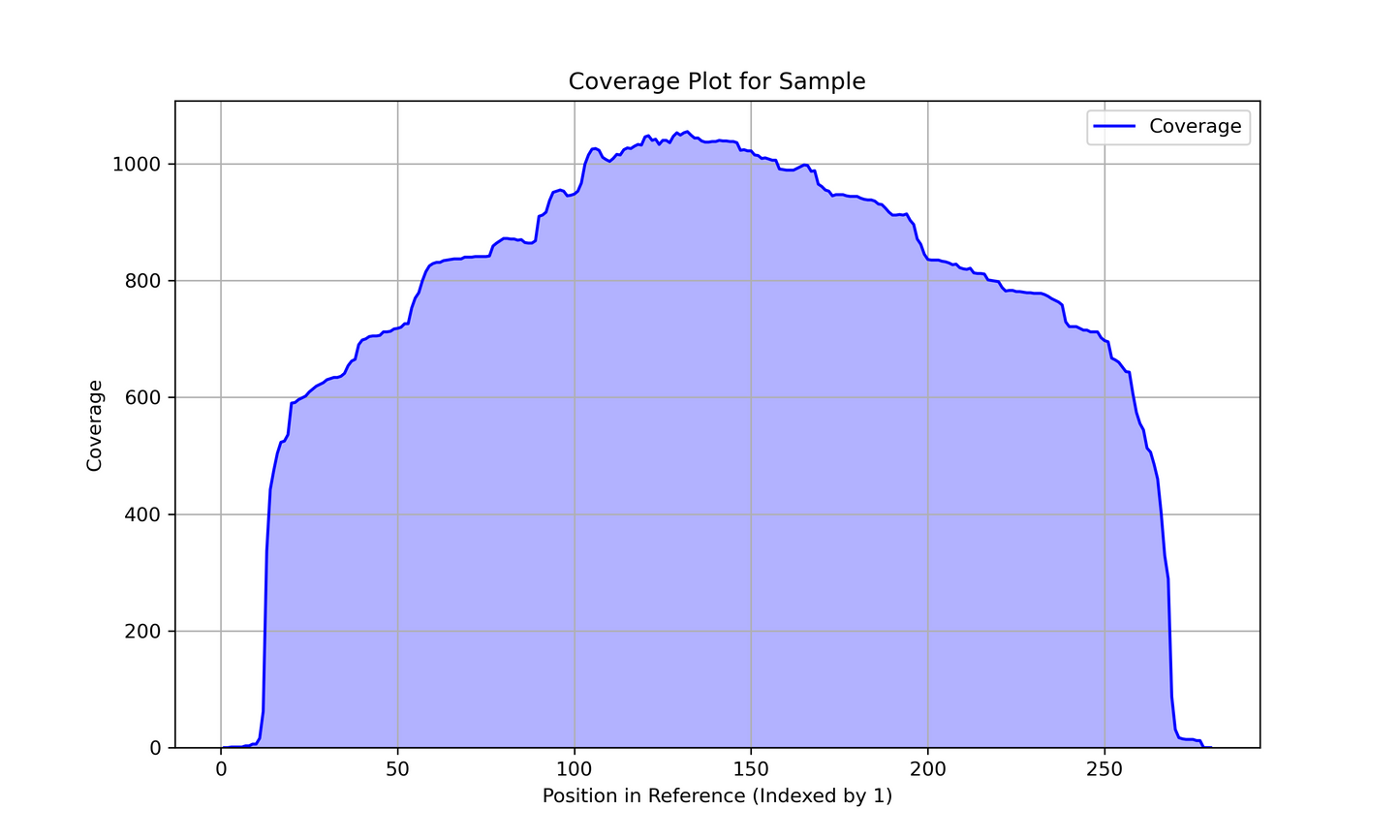
To keep our highly competitive pricing, we use the Rapid Barcode Kit (RBK) for amplicon sequencing. This does result in a small amount of trimming at the ends of the amplicons (between 15-20 bp). See the coverage example below.

In the future, we will offer a more expensive service using Ligation Sequencing Kit (LSK) to provide full length amplicons. Meanwhile, is it recommended for you to extend the PCR products so that you have a longer amplicons that still contains your desired sequence at the ends. The advantage of the RBK method is that it allows standardization for our various DNA inputs (plasmids, amplicons, and even genomes).
🚚 Sample Pickup for Bay Area customers:
We offer free Dropbox pickup on Monday to Friday
Have your orders submitted by 3:00 PM on a pickup day. Print out your confirmation email with QR code and place in the dropbox. Our courier will come between 3-4pm for pickup. Send us a request if you need a later time.
📦 Customers mailing in samples from outside the Bay Area:
Ship your samples to us via any major carrier (UPS preferred). You’ll receive a confirmation once your shipment is received.
⏱️ Turnaround Time (TAT)
Data delivery: Overnight from samples processed Monday through Friday. Data is typically ready by 8am.
📄 Data Files Returned
0.4-0.8 Megabases (Mb) of NGS data (ONT Flow Cell, R10.4.1) will be generated for each amplicon depending on the product variant you select at checkout. If you need significantly more, please contact us to discuss.
- Genbank file
- FASTA File
- .ab1 Chromatogram File
- Breseq Analysis Report
- Coverage Image
- Per base data confidence report
- Raw data (FASTQ file)
🧪 Sample Preparation Guidelines
Amplicon size: under 25 kbp.
DNA amount: 10 µL at > 20 ng/µL.
For accuracy, please measure concentration using Qubit (Nanodrop tends to be less accurate).
📂 Reference Files (optional)
If you want an alignment view:
After you place your order, you can go to seqresults.angstrominno.com to upload your references in GenBank (.gb or .gbk), Fasta (.fa or .fasta), or Snapgene format (.dna). Make sure you upload them to the corresponding Order ID for example:

If possible (not required), use the same file name for your references as the supplied sequencing sample name.