Bacterial Whole Genome Sequencing (less than 7 Mb)
Bacterial Whole Genome Sequencing (less than 7 Mb)
Please input each sample name on a separate line. Please review our sample labeling and packing instructions if this is your first order with us (especially if submitting plates).
Couldn't load pickup availability
See our demo order here: https://seqresults.angstrominno.com/orders/AI-1179
See your bacterial whole genome sequence (WGS) with automated annotations!
✅ No reference sequence is required to build the consensus.
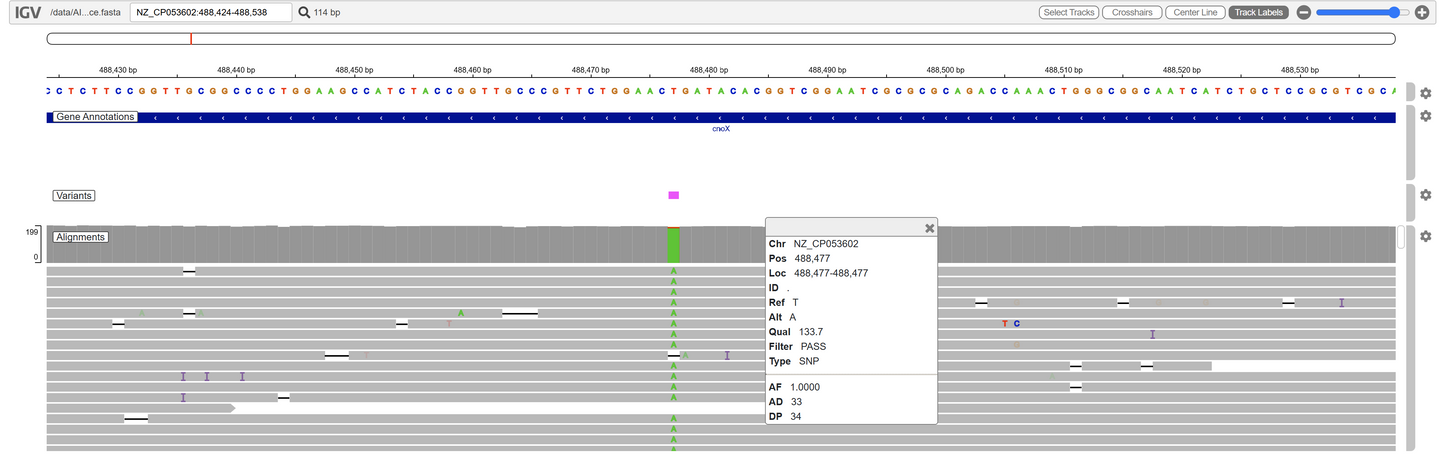
✅ If you provide a reference, we’ll align your results and highlight any mutations.
WGS samples will be processed on Tuesdays and Thursdays only.
🚚 Sample Pickup for Bay Area customers (M-F):
We offer free Dropbox pickup on Monday to Friday. Have your orders submitted by 3:00 PM on a pickup day. Print out your confirmation email with QR code and place in the dropbox. Our courier will come around 4:00 PM for pickup. Please let us know if you need later time.
📦 Customers mailing in samples from outside the Bay Area:
Ship your samples to us via any major carrier (UPS preferred).
You’ll receive a confirmation once your shipment is received.
⏱️ Turnaround Time (TAT)
Data delivery: About 1-3 days after samples have been received.
📄 Data Files Returned
About 200 Megabases (Mb) of NGS data (ONT Flow Cell, R10.4.1) will be generated for each bacterial sample.
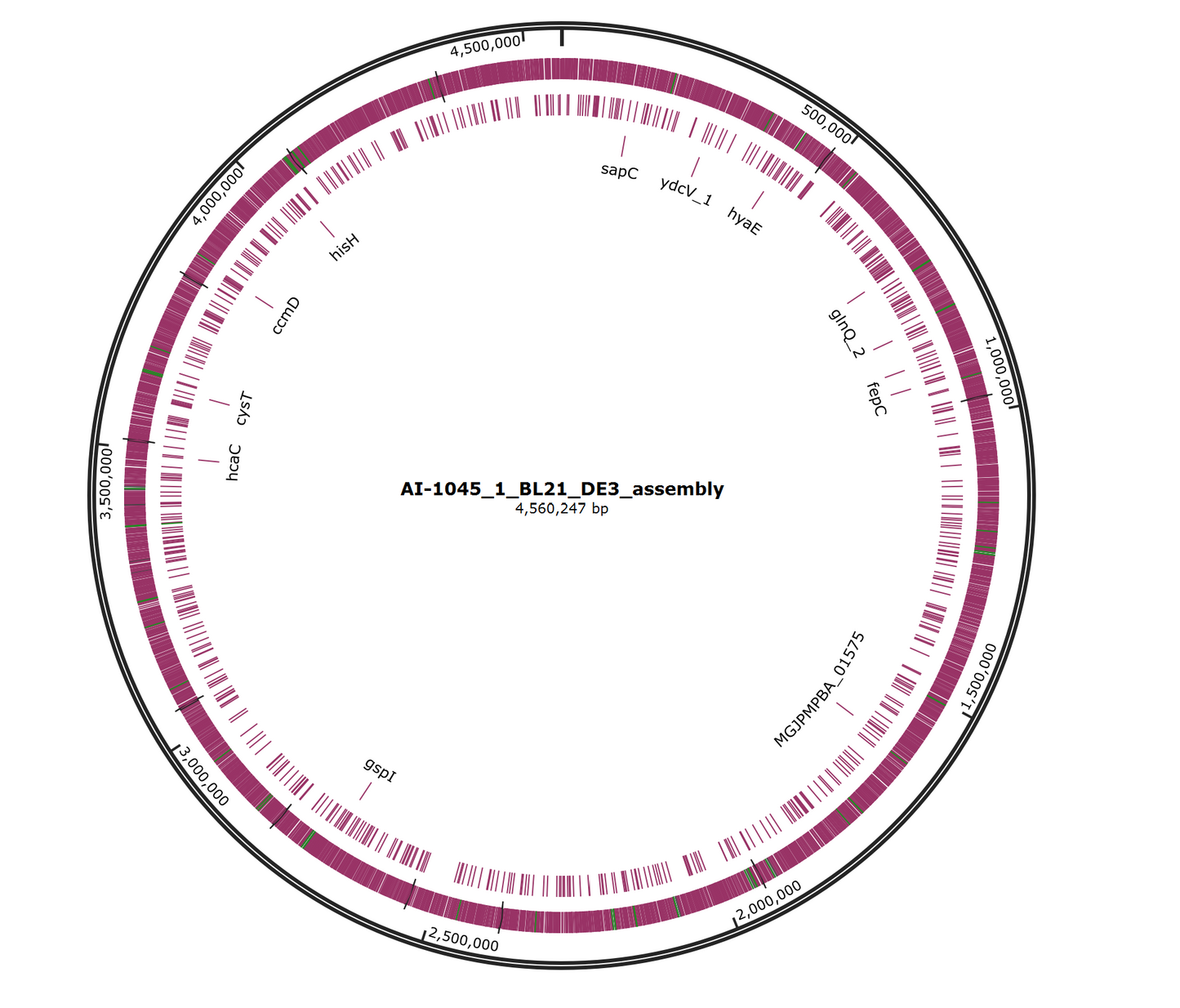
- .fasta file (de novo genome assembly)
- Properly formatted GenBank file with de novo assembly and annotations
- .gff3 file (annotations generated via Bakta using the full database)
- Assembly report with Bandage plots
- CheckM QC report (assesses completeness and contamination)
- Breseq analysis report (if reference genome is provided)*
- Raw sequencing data (.fastq.gz files)
- Interactive IGV genome browser experience (if reference genome is provided)
- Automatic determination of the closest species and reference genome
*Breseq, from the Barrick Lab (UT Austin), is usually for short reads; however, for our service, long ONT reads are truncated (≈200 bp) to fit its mapping and junction analysis. It generates clear, highly interpretable reports for mutation discovery. Your samples are still sequenced with ONT!
🧪 Sample Preparation Guidelines
DNA amount: 20 µL at > 50 ng/µL.
For accuracy, please measure concentration using Qubit (Nanodrop tends to be less accurate).
📂 Reference Files (optional)
If you want an additional alignment view:
After you place your order, you can go to seqresults.angstrominno.com to upload your references in GenBank (.gb or .gbk), Fasta (.fa or .fasta), or Snapgene format (.dna). Make sure you upload them to the corresponding Order ID for example:

If possible (not required), use the same file name for your references as the supplied sequencing sample name.